Tips & Trics for anypytools¶
Stopping and restarting simulations¶
AnyPyProcess caches the simulation results. This allows us to stop the simulations, and then later restart them again.
[1]:
from anypytools import AnyPyProcess
app = AnyPyProcess(num_processes=1)
macro = [
'load "Knee.any"',
'operation Main.MyStudy.InverseDynamics',
'run',
]
macrolist = [macro]*20
[2]:
app.start_macro(macrolist);
KeyboardInterrupt: User aborted
Completed: 18, Not processed: 2
Here we stopped the simulation using the Notebook interrupt button. Calling the start_macro() function again continues the processing and re-run any task that wasn’t completed in the first run and any task that exited with errors.
[3]:
app.start_macro(macrolist);
Completed: 20
Note: That changing the input arguments start_macro or re-instanciating the app object will erase the cache and re-run all processes.
Including meta-information in the output¶
The start_macro() also returns extra meta info, but the information is not printed by the default __repr__() function.
[4]:
from anypytools import AnyPyProcess
from anypytools.macro_commands import Load, OperationRun, Dump
app = AnyPyProcess()
macro = [
Load('Knee.any', defs={'N_STEP':10}),
OperationRun('Main.MyStudy.InverseDynamics'),
Dump('Main.MyStudy.Output.MaxMuscleActivity'),
]
result = app.start_macro(macro)[0]
result
Completed: 1
[4]:
{'Main.MyStudy.Output.MaxMuscleActivity':
array([0.00890538, 0.02510015, 0.06036529, 0.08096677, 0.08356285,
0.08356285, 0.08096678, 0.06036529, 0.02510015, 0.00890538]),
But the information is there
[5]:
result["task_macro"]
[5]:
['load "Knee.any" -def N_STEP="10"',
'operation Main.MyStudy.InverseDynamics\nrun',
'classoperation Main.MyStudy.Output.MaxMuscleActivity "Dump"',
'exit']
We can also see all task information by evaluating the result object as standard Python dictionary:
[6]:
dict(result)
[6]:
{'Main.MyStudy.Output.MaxMuscleActivity': array([0.00890538, 0.02510015, 0.06036529, 0.08096677, 0.08356285,
0.08356285, 0.08096678, 0.06036529, 0.02510015, 0.00890538]),
'task_macro_hash': '5310ca3fa2025ef0',
'task_id': 0,
'task_work_dir': 'C:\\Users\\mel\\Documents\\AnyBodySource\\AnyPyTools\\docs\\Tutorial',
'task_name': 'docs-Tutorial-0',
'task_processtime': 0.7587850093841553,
'task_macro': ['load "Knee.any" -def N_STEP="10"',
'operation Main.MyStudy.InverseDynamics\nrun',
'classoperation Main.MyStudy.Output.MaxMuscleActivity "Dump"',
'exit'],
'task_logfile': ''}
Saving output to re-process at a later time¶
The extra task meta info gives other posibilities. The results from running batch processing (i.e. output f start_macro() can be used as input to restart the same processing even if the AnyPyProcess have no cached results.
[7]:
from anypytools import AnyPyProcess
app = AnyPyProcess()
macro = [
Load('Knee.any', defs={'N_STEP':10}),
OperationRun('Main.MyStudy.InverseDynamics'),
Dump('Main.MyStudy.Output.MaxMuscleActivity'),
]
output = app.start_macro(macro)
Completed: 1
[8]:
app = AnyPyProcess()
app.start_macro(output)
Completed: 1
[8]:
[{'Main.MyStudy.Output.MaxMuscleActivity':
array([0.00890538, 0.02510015, 0.06036529, 0.08096677, 0.08356285,
0.08356285, 0.08096678, 0.06036529, 0.02510015, 0.00890538])]
The effect is that the result of an analysis can be saved to files and later restarted. The next example illustrates this.
Example: Saving data to disk while running¶
[9]:
import os
from scipy.stats import distributions
from anypytools import AnyPyProcess, AnyMacro
from anypytools.macro_commands import Load, SetValue_random, OperationRun, Dump
[10]:
tibia_knee_srel = distributions.norm([0, 0.18, 0], [0.005, 0.005, 0.005] )
femur_knee_srel = distributions.norm([0, -0.3, 0], [0.005, 0.005, 0.005] )
app = AnyPyProcess(silent=True)
mg = AnyMacro(number_of_macros = 500)
mg.extend([
Load('knee.any', defs = {'N_STEP':20}),
SetValue_random('Main.MyModel.Tibia.Knee.sRel', tibia_knee_srel),
SetValue_random('Main.MyModel.Femur.Knee.sRel', femur_knee_srel),
OperationRun('Main.MyStudy.InverseDynamics'),
Dump('Main.MyStudy.Output.MaxMuscleActivity'),
])
try:
os.remove('data.db')
except OSError:
pass
for macros in mg.create_macros_MonteCarlo(batch_size=50):
app.start_macro(macros)
app.save_results('data.db', append=True)
print('Data saved')
print('Done')
Data saved
Data saved
Data saved
Data saved
Data saved
Data saved
Data saved
Data saved
Data saved
Data saved
Done
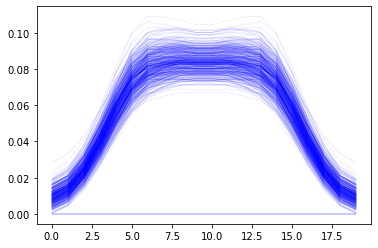
All this stored data can be be reloaded
[11]:
reloaded_results = app.load_results('data.db')
print('Entries in file: {}'.format(len(reloaded_results)))
Entries in file: 500
[12]:
reloaded_results[456:457]
[12]:
[{'Main.MyStudy.Output.MaxMuscleActivity':
array([0.0145094 , 0.01838543, 0.02723008, 0.03983322, 0.05388536,
0.06596098, 0.07287488, 0.07568929, 0.07757901, 0.07764066,
0.07764066, 0.07757901, 0.07568929, 0.07287487, 0.06596096,
0.05388536, 0.03983322, 0.02723023, 0.01838528, 0.0145094 ])]
[13]:
%matplotlib inline
import matplotlib.pyplot as plt
plt.plot(reloaded_results['MaxMuscleAct'].T, 'b', lw=0.2, alpha = 0.3);
[ ]:
