Advanced Examples#
Making a Monte Carlo parameter study#
In this example, we will make a Monte Carlo study. This is a case where the python library shows it’s advantage.
We will make a Monte Carlo study on the position of patella tendon insertion and origin in the simplified knee model used in the first tutorial. Thus, we need some macros that change two sRel variables in the model. In this case, we choose the values from a truncated normal distribution, but any statistical distribution could have been used.
from scipy.stats import distributions
# Truncated normal between +/- 2SD.
patella_tendon_insertion = distributions.truncnorm(-2,2,[0.02, 0.12, 0], [0.01,0.01,0.01])
patella_tendon_origin = distributions.truncnorm(-2,2,[0.0,-0.03, 0], [0.01,0.01,0.01])
from anypytools import macro_commands as mc
macro = [
mc.SetValue_random('Main.MyModel.Tibia.Patella2.sRel', patella_tendon_insertion),
mc.SetValue_random('Main.MyModel.Patella.Lig.sRel', patella_tendon_origin)
]
macro
[classoperation Main.MyModel.Tibia.Patella2.sRel "Set Value" --value="{0.02,0.12,2.78291642467e-18}",
classoperation Main.MyModel.Patella.Lig.sRel "Set Value" --value="{2.78291642467e-18,-0.03,2.78291642467e-18}"]
The default representation of the macro is 50% percentile values from the distribution (i.e. the mean value). To generate something random we need the AnyMacro helper class.
The AnyMacro helper class can generate multiple macros from a single macro.
from anypytools import AnyMacro
mg = AnyMacro(macro)
monte_carlo_macros = mg.create_macros_MonteCarlo(100)
The first generated macro just has the default values.
monte_carlo_macros[0]
['classoperation Main.MyModel.Tibia.Patella2.sRel "Set Value" --value="{0.02,0.12,2.78291642467e-18}"',
'classoperation Main.MyModel.Patella.Lig.sRel "Set Value" --value="{2.78291642467e-18,-0.03,2.78291642467e-18}"']
The next two macros have random offsets.
monte_carlo_macros[1:4]
[['classoperation Main.MyModel.Tibia.Patella2.sRel "Set Value" --value="{0.0144267872987,0.109982579448,-0.01776142425}"',
'classoperation Main.MyModel.Patella.Lig.sRel "Set Value" --value="{0.00182301737147,-0.0385242952422,0.0183692935384}"'],
['classoperation Main.MyModel.Tibia.Patella2.sRel "Set Value" --value="{0.0242051783405,0.103809162986,0.00739209161022}"',
'classoperation Main.MyModel.Patella.Lig.sRel "Set Value" --value="{-0.00636881243957,-0.0180115562062,0.0118074789971}"'],
['classoperation Main.MyModel.Tibia.Patella2.sRel "Set Value" --value="{0.0249230579594,0.115461116587,-0.00698883214514}"',
'classoperation Main.MyModel.Patella.Lig.sRel "Set Value" --value="{0.00483548485266,-0.014494203176,0.00744105538977}"']]
Now let us expand the macro to also load and run the model.
macro = [
mc.Load('Knee.any'),
mc.SetValue_random('Main.MyModel.Tibia.Patella2.sRel', patella_tendon_insertion ) ,
mc.SetValue_random('Main.MyModel.Patella.Lig.sRel', patella_tendon_origin ) ,
mc.RunOperation('Main.MyStudy.InverseDynamics'),
mc.Export('Main.MyStudy.Output.Abscissa.t', "time"),
mc.Export('Main.MyStudy.Output.MaxMuscleActivity', "MaxMuscleActivity")
]
mg = AnyMacro(macro, seed=1)
monte_carlo_macros = mg.create_macros_MonteCarlo(100)
Running the Monte Carlo macro#
The macro is passed to the AnyPyProcess object which excutes the macros
from anypytools import AnyPyProcess
app = AnyPyProcess()
monte_carlo_results = app.start_macro( monte_carlo_macros )
Completed: 100
The output object (monte_carlo_result) is a list-like object where each element is a dictionary with the output of the corresponding simulation.
print('Length :', len(monte_carlo_results) )
print('Data keys in first element: ', list(monte_carlo_results[0].keys()))
Length : 100
Data keys in first element: ['time', 'MaxMuscleActivity', 'task_macro_hash', 'task_id', 'task_work_dir', 'task_name', 'task_processtime', 'task_macro', 'task_logfile']
Filtering results with Errors#
If any model errors occured in some of the simulations, then the output may be missing. That can be a problem for futher processing. So we may want to remove those simulations.
That is easily done. Results from simuations with errors will contain a an ‘ERROR’ key. We can use that to filter the results.
monte_carlo_results[:] = [
output
for output in monte_carlo_results
if 'ERROR' not in output
]
Extracting data from the resutls#
The object looks and behaves like a list, but it can do more things than a standard Python list. If we use it as a dictionary and pass the variable names, it will return that variable concatenated over all runs.
monte_carlo_results['MaxMuscleActivity']
array([[0.00890538, 0.00927552, 0.00986515, ..., 0.00986515, 0.00927552,
0.00890538],
[0.00761831, 0.00793615, 0.00844398, ..., 0.00844398, 0.00793614,
0.00761831],
[0.00965061, 0.01004897, 0.0106812 , ..., 0.0106812 , 0.01004897,
0.00965062],
...,
[0.01007754, 0.0104962 , 0.01116247, ..., 0.01116247, 0.01049619,
0.01007754],
[0.01026166, 0.01068757, 0.01136452, ..., 0.01136452, 0.01068757,
0.01026167],
[0.00981378, 0.01021894, 0.01086207, ..., 0.01086207, 0.01021893,
0.00981379]], shape=(100, 100))
Plotting the results#
Finally we can plot the result of the Monte Carlo study
%matplotlib inline
import matplotlib.pyplot as plt
time = monte_carlo_results['time'][0]
max_muscle_activity = monte_carlo_results["MaxMuscleActivity"]
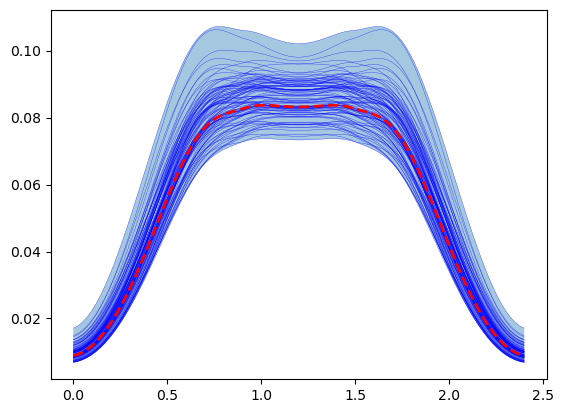
plt.fill_between(time, max_muscle_activity.min(0), max_muscle_activity.max(0),alpha=0.4 )
for trace in max_muscle_activity:
plt.plot(time, trace,'b', lw=0.2 )
# Plot result with the mean of the inputs ( stored in the first run)
plt.plot(time, max_muscle_activity[0],'r--', lw = 2, )
[<matplotlib.lines.Line2D at 0x22124a60830>]
Latin Hypercube sampling#
Monte Carlo studies are not very efficient when investigating the effect of many parameters. It quickly becomes necessary to run the model thousands of times. Not very convenient if the AnyBody model takes a long time to run.
Another approach is to use Latin Hypercube sampling. From Wikipedia
Latin hypercube sampling (LHS) is a statistical method for generating a sample of plausible collections of parameter values from a multidimensional distribution. The sampling method is often used to construct computer experiments.
Using LHS we can generate a sample that better spans the whole multidimensional space. Thus, fever model evaluations are necessary. See pyDOE for examples (and explanation of the criterion/iterations parameters which can be parsed to create_macros_LHS()).
To following uses LHS to do the same as in the previous example:
patella_tendon_insertion = distributions.norm([0.02, 0.12, 0], [0.01,0.01,0.01])
patella_tendon_origin = distributions.norm([0.0,-0.03, 0], [0.01,0.01,0.01])
macro = [
mc.Load('Knee.any'),
mc.SetValue_random('Main.MyModel.Tibia.Patella2.sRel', patella_tendon_insertion ) ,
mc.SetValue_random('Main.MyModel.Patella.Lig.sRel', patella_tendon_origin ) ,
mc.RunOperation('Main.MyStudy.InverseDynamics'),
mc.Export('Main.MyStudy.Output.Abscissa.t', 'time'),
mc.Export('Main.MyStudy.Output.MaxMuscleActivity', 'MaxMuscleActivity')
]
mg = AnyMacro(macro, seed=1)
LHS_macros = mg.create_macros_LHS(25)
D:\repos\AnyPyTools\.pixi\envs\jupyter\Lib\site-packages\pyDOE\doe_factorial.py:192: SyntaxWarning: "\-" is an invalid escape sequence. Such sequences will not work in the future. Did you mean "\\-"? A raw string is also an option.
A = [item for item in re.split('\-?\s?\+?', gen) if item] # remove empty strings
from anypytools import AnyPyProcess
app = AnyPyProcess()
lhs_results = app.start_macro(LHS_macros)
Completed: 25
%matplotlib inline
import matplotlib.pyplot as plt
time = lhs_results['Abscissa.t'][0]
max_muscle_act = lhs_results['MaxMuscleActivity']
plt.fill_between(time, max_muscle_act.min(0), max_muscle_act.max(0), alpha=0.4)
for trace in max_muscle_act:
plt.plot(time, trace,'b', lw=0.2 )
# Plot the mean value that was stored in the first results
plt.plot(time, max_muscle_act.mean(0),'r--', lw = 2, )
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[14], line 4
1 get_ipython().run_line_magic('matplotlib', 'inline')
2 import matplotlib.pyplot as plt
----> 4 time = lhs_results['Abscissa.t'][0]
5 max_muscle_act = lhs_results['MaxMuscleActivity']
6 plt.fill_between(time, max_muscle_act.min(0), max_muscle_act.max(0), alpha=0.4)
File D:\repos\AnyPyTools\anypytools\tools.py:400, in AnyPyProcessOutputList.__getitem__(self, item)
396 key_in_all_elements = all(
397 super(AnyPyProcessOutput, e).__contains__(key) for e in self.list
398 )
399 if not key_in_all_elements:
--> 400 raise KeyError(
401 f" The key: '{key}' is not present in all elements of the output."
402 ) from None
403 try:
404 data = np.array(
405 [super(AnyPyProcessOutput, e).__getitem__(key) for e in self.list]
406 )
KeyError: " The key: 'Abscissa.t' is not present in all elements of the output."
